Nicodem
Super Moderator
      
Posts: 4230
Registered: 28-12-2004
Member Is Offline
Mood: No Mood
|
|
bbcode support for SMILES structures to images
I wish we could implement a SMILES module to show chemical structures. That would be much more useful. The defunct Hive forum had one that was very
good. I have no idea what it would take, or if it is possible at all on this platform. There are some free SMILES, CAS or chemical name to structure
translators on the web (for example: http://www.openmolecules.org/name2structure.html). I wander if it is possible to implement their service into the forum code?
…there is a human touch of the cultist “believer” in every theorist that he must struggle against as being
unworthy of the scientist. Some of the greatest men of science have publicly repudiated a theory which earlier they hotly defended. In this lies their
scientific temper, not in the scientific defense of the theory. - Weston La Barre (Ghost Dance, 1972)
Read the The ScienceMadness Guidelines!
|
|
|
Polverone
Now celebrating 21 years of madness
        
Posts: 3186
Registered: 19-5-2002
Location: The Sunny Pacific Northwest
Member Is Offline
Mood: Waiting for spring
|
|
It was a lot more work than MathJax but I integrated the openmolecules.net SMILES rendering service. I turned bbcode off for this post to show the
tags.
Fructose:
[smiles]O[C@H]1[C@H](O)[C@H](O[C@]1(O)CO)CO[/smiles]
Caffeine:
[smiles]Cn1cnc2c1c(=O)n(c(=O)n2C)C[/smiles]
EDIT: Apparently the name service recognizes more than SMILES!
Water:
[smiles]water[/smiles]
PETN:
[smiles]PETN[/smiles]
[Edited on 7-13-2014 by Polverone]
PGP Key and corresponding e-mail address
|
|
|
Polverone
Now celebrating 21 years of madness
        
Posts: 3186
Registered: 19-5-2002
Location: The Sunny Pacific Northwest
Member Is Offline
Mood: Waiting for spring
|
|
And now the real result:
Fructose:

Caffeine:
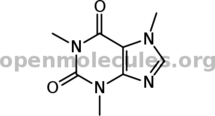
EDIT: Apparently the name service recognizes more than SMILES!
Water:
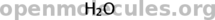
PETN:

[Edited on 7-13-2014 by Polverone]
PGP Key and corresponding e-mail address
|
|
|
Polverone
Now celebrating 21 years of madness
        
Posts: 3186
Registered: 19-5-2002
Location: The Sunny Pacific Northwest
Member Is Offline
Mood: Waiting for spring
|
|
And finally, the meat of the code. To integrate it in another message board you'd want to instantiate a SmilesCode instance and call the
bbcode_replace method on the message body after other bbcode tags have been transformed.
| Code: |
<?php
/**
* SMILES image manager based on openmolecules.org name2structure
* service.
*
**/
if (!defined('IN_CODE')) {
header('HTTP/1.0 403 Forbidden');
exit("Not allowed to run this file directly.");
}
define("IMAGE_DIR", "/var/www/smiles/");
define("SMILES_IMG_LOC", "www.sciencemadness.org/smiles/");
define("OM_PREFIX", "http://n2s.openmolecules.org/?name=");
define("FAILURE_IMAGE", "smilesfailure.png");
class SmilesCode {
// convert SMILES to img representing corresponding structure
public function render($smiles) {
$smiles = trim($smiles);
$key = md5($smiles);
$image_name = $key . '.png';
$file_name = IMAGE_DIR . $image_name;
// image not stored locally -- try to generate image file
if (!file_exists($file_name)) {
$file_name = $this->generate_file($smiles, $image_name);
}
if (!empty($_SERVER['HTTPS']) && $_SERVER['HTTPS'] !== 'off') {
$protocol = 'https://';
}
else {
$protocol = 'http://';
}
$img_url = $protocol . SMILES_IMG_LOC . $image_name;
$img = "<img src=\"$img_url\"></img>";
return $img;
}
// generate an image file using the openmolecules.net server
public function generate_file($smiles, $image_name) {
$remote_url = OM_PREFIX . $smiles;
$http = curl_init($remote_url);
curl_setopt($http, CURLOPT_RETURNTRANSFER, 1);
$result = curl_exec($http);
$status = curl_getinfo($http, CURLINFO_HTTP_CODE);
curl_close($http);
if ($status != 200) {
$image_name = FAILURE_IMAGE;
}
else {
$out_name = IMAGE_DIR . $image_name;
file_put_contents($out_name, $result);
}
return $image_name;
}
// replace all bbcode SMILES with molecular images
public function bbcode_replace($message) {
$codes = $this->extract_smiles($message);
foreach ($codes as $code) {
$bare_smiles = str_replace(array('[smiles]', '[/smiles]'),
array(), $code);
$rendered = $this->render($bare_smiles);
$message = str_replace($code, $rendered, $message);
}
return $message;
}
// get all bbcode SMILES markup
public function extract_smiles($message) {
$codes = array();
$offset = 0;
$begin_tag = '[smiles]';
$end_tag = '[/smiles]';
$current_tag = $begin_tag;
while ($offset < strlen($message)) {
$old_pos = $offset;
$pos = strpos($message, $current_tag, $offset);
if ($pos === false) {
break;
}
else {
$offset = $pos;
}
// found begin -- switch to end search
if ($current_tag == $begin_tag) {
$current_tag = $end_tag;
}
// found end -- capture contents and switch back to begin search
else {
$smile = substr($message, $old_pos,
$pos - $old_pos + strlen($end_tag));
array_push($codes, $smile);
$current_tag = $begin_tag;
}
}
return $codes;
}
}
?>
|
[Edited on 7-13-2014 by Polverone]
PGP Key and corresponding e-mail address
|
|
|
Brain&Force
Hazard to Lanthanides
    
Posts: 1302
Registered: 13-11-2013
Location: UW-Madison
Member Is Offline
Mood: Incommensurately modulated
|
|
OK, I can't resolve this SMILES input:
[I-].[I-].[I-].[Tb+3](~O=C1C=C(C)N(C)N1C:2:C:C:C:C:C:2)(~O=C3C=C(C)N(C)N3C:4:C:C:C:C:C:4)(~O=C5C=C(C)N(C)N5C:6:C:C:C:C:C:6)(~O=C7C=C(C)N(C)N7C:8:C:C:C
:C:C:8)(~O=C9C=C(C)N(C)N9C:%10:C:C:C:C:C:%10)~O=C%11C=C(C)N(C)N%11C:%12:C:C:C:C:C:%12
This is hexakis(antipyrine)terbium iodide - a coordination complex with coordination bonds.
Diamminesilver(I) doesn't work either:
[Ag+].N.N
Tetrachloronickelate works.
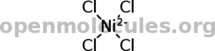
Is there something wrong with the input, or is this a problem on the software's end? The first structure was generated by me, the last two were pulled
off of ChemSpider.
At the end of the day, simulating atoms doesn't beat working with the real things...
|
|
|
Polverone
Now celebrating 21 years of madness
        
Posts: 3186
Registered: 19-5-2002
Location: The Sunny Pacific Northwest
Member Is Offline
Mood: Waiting for spring
|
|
There is a stray space in your SMILES: "...N7C:8:C:C:C :C:C:8..."
Even with the space removed, the openmolecules.net service cannot properly parse it. In fact, out of the toolkits incorporated into Cinfony, only
RDKit could parse it: http://www.rdkit.org/
It looks like most chemoinformatics toolkits are not built with coordination chemistry in mind.
I had considered using RDKit to write the back end of the SMILES rendering service for here on the board. But I tested it with some complex chiral
organics and the openmolecules.net service made better images, plus it would be more complicated to wrap RDKit. I suppose I could add an RDKit
fallback renderer to try to handle whatever openmolecules.org fails on.
PGP Key and corresponding e-mail address
|
|
|
HeYBrO
Hazard to Others
  
Posts: 289
Registered: 6-12-2013
Location: 'straya
Member Is Offline
Mood: 
|
|
I Know who has a new signature 
EdiT: it is way too big for a signature, so maybe disable that...
[Edited on 13-7-2014 by HeYBrO]
[Edited on 13-7-2014 by HeYBrO]

|
|
|
Polverone
Now celebrating 21 years of madness
        
Posts: 3186
Registered: 19-5-2002
Location: The Sunny Pacific Northwest
Member Is Offline
Mood: Waiting for spring
|
|
I added code to automatically trim excess border space from generated images. They're still kind of large though. Should I use thumbnails plus links
to full size images?
PGP Key and corresponding e-mail address
|
|
|
Nicodem
Super Moderator
      
Posts: 4230
Registered: 28-12-2004
Member Is Offline
Mood: No Mood
|
|
Beautiful job! The openmolecules.org service has a few limitations, but at least it is free and it looks like it has ambitions to stay active for the
future. I agree that the images are big, but I would not like the idea of the thumbnails. Is it possible to rather automatically resize them? I think
they would still be OK at half size.
Also note that the service recognizes not just SMILES, chemical names, some trivial names and generic abbreviations, it also takes CAS numbers.
For example (with codes switched off):
trivial name: strychnine
[smiles]strychnine[/smiles]
CAS number: ergocristine = 511-08-0
[smiles]511-08-0[/smiles]
generic API name: indinavir
[smiles]indinavir[/smiles]
|
|
|
Nicodem
Super Moderator
      
Posts: 4230
Registered: 28-12-2004
Member Is Offline
Mood: No Mood
|
|
...resulting in...
trivial name: strychnine
CAS number: ergocristine = 511-08-0

generic API name: indinavir

…there is a human touch of the cultist “believer” in every theorist that he must struggle against as being
unworthy of the scientist. Some of the greatest men of science have publicly repudiated a theory which earlier they hotly defended. In this lies their
scientific temper, not in the scientific defense of the theory. - Weston La Barre (Ghost Dance, 1972)
Read the The ScienceMadness Guidelines!
|
|
|
Chemosynthesis
International Hazard
    
Posts: 1071
Registered: 26-9-2013
Member Is Offline
Mood: No Mood
|
|


Going to have to work on my salt SMILES. The wiki didn't work.
[Edited on 14-7-2014 by Chemosynthesis]
Just saw that  . .
Hmm. 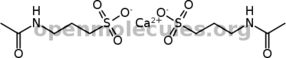
Thank you both!
[Edited on 14-7-2014 by Chemosynthesis]
|
|
|
Polverone
Now celebrating 21 years of madness
        
Posts: 3186
Registered: 19-5-2002
Location: The Sunny Pacific Northwest
Member Is Offline
Mood: Waiting for spring
|
|
They're automatically scaled to half size now. Too blurry?
Edit: Chemosynthesis, the smiles tag should be in lower case. I didn't try to make it case-insensitive.
[Edited on 7-14-2014 by Polverone]
PGP Key and corresponding e-mail address
|
|
|
Chemosynthesis
International Hazard
    
Posts: 1071
Registered: 26-9-2013
Member Is Offline
Mood: No Mood
|
|
I might prefer 0.75 scale for easier visibility on valences, if that sounds good to others.
|
|
|
Nicodem
Super Moderator
      
Posts: 4230
Registered: 28-12-2004
Member Is Offline
Mood: No Mood
|
|
They look perfectly fine at half size to me, but I guess that pretty much depends on the monitor size and settings at the user end.
Chemosynthesis, the molecules I posted above are relatively larger than anything we usually discuss on this forum. Check the "normal sized" compound
like the ones that Polverone posted above. They should be fine unless you have a small monitor or screen resolution settings.
The size is still slightly too big to depict simple schemes as one liners (unless I reduce the browser display size), but it is comprehensible with
some effort:
[smiles]c1(cc(c(cc1)O)OC)C=O[/smiles][b][size=6]+[/size][/b][smiles]acetophenone[/smiles] [b][size=6]→[/size][/b]
[smiles]c1(cc(c(cc1)O)OC)/C=C/C(c2ccccc2)=O[/smiles]
Note: For the HTML codes of various arrows useful in chemical equations, see the list at
[url=http://character-code.com/arrows-html-codes.php]http://character-code.com/arrows-html-codes.php[/url]
|
|
|
Nicodem
Super Moderator
      
Posts: 4230
Registered: 28-12-2004
Member Is Offline
Mood: No Mood
|
|
...resulting in...
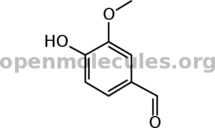 + + → → 
Note: For the HTML codes of various arrows useful in chemical equations, see the list at http://character-code.com/arrows-html-codes.php
…there is a human touch of the cultist “believer” in every theorist that he must struggle against as being
unworthy of the scientist. Some of the greatest men of science have publicly repudiated a theory which earlier they hotly defended. In this lies their
scientific temper, not in the scientific defense of the theory. - Weston La Barre (Ghost Dance, 1972)
Read the The ScienceMadness Guidelines!
|
|
|
TheChemiKid
Hazard to Others
  
Posts: 493
Registered: 5-8-2013
Location: ̿̿ ̿̿ ̿'̿'̵͇̿̿з=༼ ▀̿̿Ĺ̯̿̿▀̿ ̿ ༽
Member Is Offline
Mood: No Mood
|
|
Test for vanillin:

EDIT: Hmm, the wiki picture looks like this, so it confused me. Sorry for the double post.

[Edited on 7-14-2014 by TheChemiKid]
When the police come
\( * O * )/ ̿̿ ̿̿ ̿'̿'̵͇̿̿з=༼ ▀̿̿Ĺ̯̿̿▀̿ ̿ ༽
|
|
|
Chemosynthesis
International Hazard
    
Posts: 1071
Registered: 26-9-2013
Member Is Offline
Mood: No Mood
|
|
Quote: Originally posted by Nicodem  |
Chemosynthesis, the molecules I posted above are relatively larger than anything we usually discuss on this forum. Check the "normal sized" compound
like the ones that Polverone posted above. They should be fine unless you have a small monitor or screen resolution settings. |
Checked on a bigger screen and they look good. I tend to forget that I often use small screens for space, or my phone.
Awesome work again!
|
|
|
arkoma
Redneck Overlord
      
Posts: 1763
Registered: 3-2-2014
Location: On a Big Blue Marble hurtling through space
Member Is Offline
Mood: украї́нська
|
|
This is wonderful--Kudos!

| Code: | one of my favorite anthocyanidins, and all I had to
enter was [smiles]petunidin[/smiles] Great job |
"We believe the knowledge and cultural heritage of mankind should be accessible to all people around the world, regardless of their wealth, social
status, nationality, citizenship, etc" z-lib
|
|
|
The Volatile Chemist
International Hazard
    
Posts: 1981
Registered: 22-3-2014
Location: 'Stil' in the lab...
Member Is Offline
Mood: Copious
|
|
Cheers! This is a great blessing! Thanks for the work put into it, I've always loved SMILES!
|
|
|
The Volatile Chemist
International Hazard
    
Posts: 1981
Registered: 22-3-2014
Location: 'Stil' in the lab...
Member Is Offline
Mood: Copious
|
|
If you don't mind, I'm gonna test it here...

[Edited on 7-20-2014 by The Volatile Chemist]
That second one's Cephalostatin-1
It really doesn't like it... I think it has something to do with the '%' character for binding when the binding number is > 9, but I don't know...
According to wikipedia, it's smiles should be | Code: | C[C@@](C)(O1)C[C@@H](O)[C@@]1(O2)[C@@H](C)[C@@H]3CC=C4[C@]3(C2)C(=O)C[C@H]5[C@H]4CC[C@@H](C6)[C@]5(C)Cc(n7)c6nc(C[C@@]89(C))c7C[C@@H]8CC[C@@H]%10[C@@H]9C[C@@H](O)[C@@]%11(C)C%10=C[C@H](O%12)[C@]%11(O)[C@H](C)[C@]%12(O%13)[C@H](O)C[C@@]%13(C)CO |
[Edited on 7-20-2014 by The Volatile Chemist]
[Edited on 7-20-2014 by The Volatile Chemist]
|
|
|