bismuth89 - 1-12-2011 at 22:34
This is the first time I deal with MS data as I'm a student in pharmacy doing my thesis in the organic chemistry lab of my university, so please help
me, thanks a lot.
I'm trying to synthesize T3 (triiodothyronine) from tyrosine, and after 3 reactions I suppose to obtain this compound:

http://pubchem.ncbi.nlm.nih.gov/summary/summary.cgi?cid=5387...
MW= 341.2735g/mol
I send a sample of the compound to a center to perform LC-MS (as my lab doesn't have MS equipment), and they send back the spectra for me with email:

As you can see, the peak with m/z= 338.8 is the highest peak, it's about M-2 compare to my expecting compound.
All the information about the LC-MS process I can have is present in the spectra above (name of instrument...)
So can I conclude that the compound I synthesized is the one I expected? Why is it M-2?
Please help me with this as I'm really confused now and I've synthesized a fairly large amount of this compound.
I'd really appreciate your help.
[Edited on 2-12-2011 by bismuth89]
[Edited on 2-12-2011 by bismuth89]
DJF90 - 2-12-2011 at 01:14
As its a neg mass spec, you're expecting RMM - H as your parent peak. In this case, it looks as if the dianion may predominantly form; that phenol OH
and either amide NH or α-proton of ester look like good candidates. However, I am only familiar with electrospray MS.
bismuth89 - 2-12-2011 at 02:41
Thanks for your help, when I use chembiodraw to calculate the MW, it says the exact mass is 341.09
If the molecule lost 2H then the fragment's mass should be 339.07, while in the spectra it's 338.8.
Should both numbers be rounded to 339, or there's a problem?
fledarmus - 2-12-2011 at 05:07
Mass specs are actually measuring the ratio of mass to charge of an ion - m/z. If you have a dianion, you would expect the mass spec to show something
around 169 rather than 339.
I suspect that when you pull the proton from the hydroxy to form the oxygen anion, those electrons go back to the ring to form a ketone, eliminating a
neutral hydrogen atom alpha to the benzene ring in the para position and forming a double bond in the para position (a quinone-like structure - sorry
for the poor description, I'm not good with inserting images). Essentially, your fragmentation is generating an (M-2)- ion, rather than your original
(M-1)- ion.
ScienceSquirrel - 2-12-2011 at 05:34
Your best bet would be to get an NMR.
It's not it
Arrhenius - 5-12-2011 at 08:37
Hi Bismuth,
I'm going to take a wild guess that your three steps are:
1.) nitration
2.) esterification
3.) N-acetylation
Perhaps not in that order, but probably.
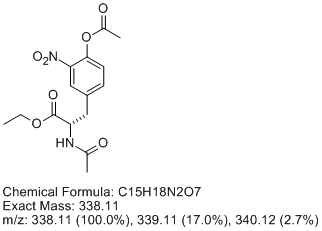
Be critical of data - always! I can't tell whether its negative or positive ionization. Here's what I think you've got, resulting from incomplete
nitration (totally reasonable, I've done this reaction and unless you beat on it, it only goes once) and overacetylation (also easy to do, the
nitrophenol is quite acidic). Let me know what you think. The data is perfectly consistent with [M+H]+ = 339, if this is a positive ion MS, which I
suspect it is.
[Edited on 6-12-2011 by Arrhenius]
UnintentionalChaos - 5-12-2011 at 09:35
A 1H NMR would resolve this very fast. If you get a 2H singlet in the aromatic region, congrats, it's dinitrated. If you get a 1H singlet and two 1H
doublets, Arrhenius is onto something.